ASARS–CoV–2 lineage A variant (A.23.1) with altered spike has emerged and is dominating the current Uganda epidemic and has spread to 25 other countries.

The Mutated covid19 virus known as A.23.1 virus sequences have 4 or 5 amino acid changes in the spike protein (now defining lineage A.23.1) plus additional protein changes in nsp3, nsp6, ORF8 and ORF9. The P681R spike change encoded by all recent genomes adds a basic amino acid adjacent to the spike furin cleavage site. This same change has been shown in vitro to enhance the fusion activity of the SARS-CoV-2 spike protein, likely due to increased cleavage by the cellular furin protease importantly, a similar change (P681H) is encoded by the recently emerging VOC B.1.1.7 that is now spreading globally across 75 countries.
There are also changes in the spike N-terminal domain (NTD), a known target of immune selection, observed in samples, including P26S and R102I plus 8 additional singleton changes (observed in only one genome.
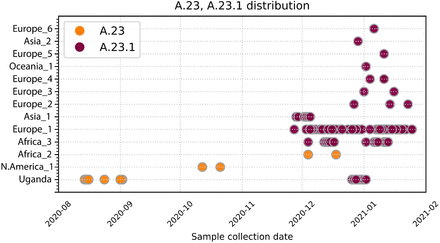
All available SARS-CoV-2 complete genomes annotated as complete and lineage A from GISAID were retrieved on Feb 4 2021 and lineage typed using Pangolin(27). and confirmed as A.23 and A.23.1 by extracting examining the encoded spike protein. A.23 and A.23.1 genomes were plotted by country and sample collection date. Countries were anonymized by continent.

The viruses detected in Amuru and Kitgum met the criteria for a new SARS-CoV-2 lineage by clustering together on a global phylogenetic tree, sharing epidemiological history and source from a single geographical origin, and encoding multiple defining SNPs. These features including especially the three spike changes F157L, Q613H and V367F define the new lineage A.23. Continued circulation and evolution of A.23 in Uganda was observed and two additional changes in spike R102I and P681R were observed in December 2020 in Kampala; these SNPs define the sub-lineage A.23.1. Additional changes in non-spike regions also define the A.23 and A.23.1, including nsp3: E95K, nsp6: M86I, L98F, ORF 8: L84S, E92K and ORF9 N: S202N, Q418H. These new lineages can be assigned since pangolin version v2.1.10 and pangoLEARN data release 2021-02-01. Screening SARS-CoV-2 genomic data from GISAID, viruses from A.23 and A.23.1 are now found in 12 countries outside Uganda (from Africa, Asia, Europe, North America and Oceania) indicating global movement of the newly emerging variants Outside of Uganda, A.23.1 was found in another country in Africa (Africa_3) from the end of November in 9 different countries across Europe (6 countries), Asia (2 countries) and Oceania (1 country). Of note, the international flights out of Uganda were restarted on 1 October 2020 with frequent flights to Europe and US overlaying via a country in Africa or Asia. Phylogenetic analysis supports the close evolution of A.23 to A.23.1 We report the emergence and spread of a new SARS-CoV-2 variant of the A lineage (A.23.1) with multiple protein changes throughout the viral genome. We suspect that all the emerging SARS-CoV-2 may be adjusting to infection and replication in humans and it is notable that the VOCs and lineage A.23.1 share a common patterns in their evolution. The spike changes are best understood due to the massive global effort to define the receptor and develop vaccines against the infection. The analysis reveals common functions of SARS-CoV-2 that have been altered in all four variants, especially nsp6 and the ORF 7, 8 and 9.
Read it for yourself click the link below
ASARS-CoV-2 lineage A variant (A.23.1) with altered spike has emerged